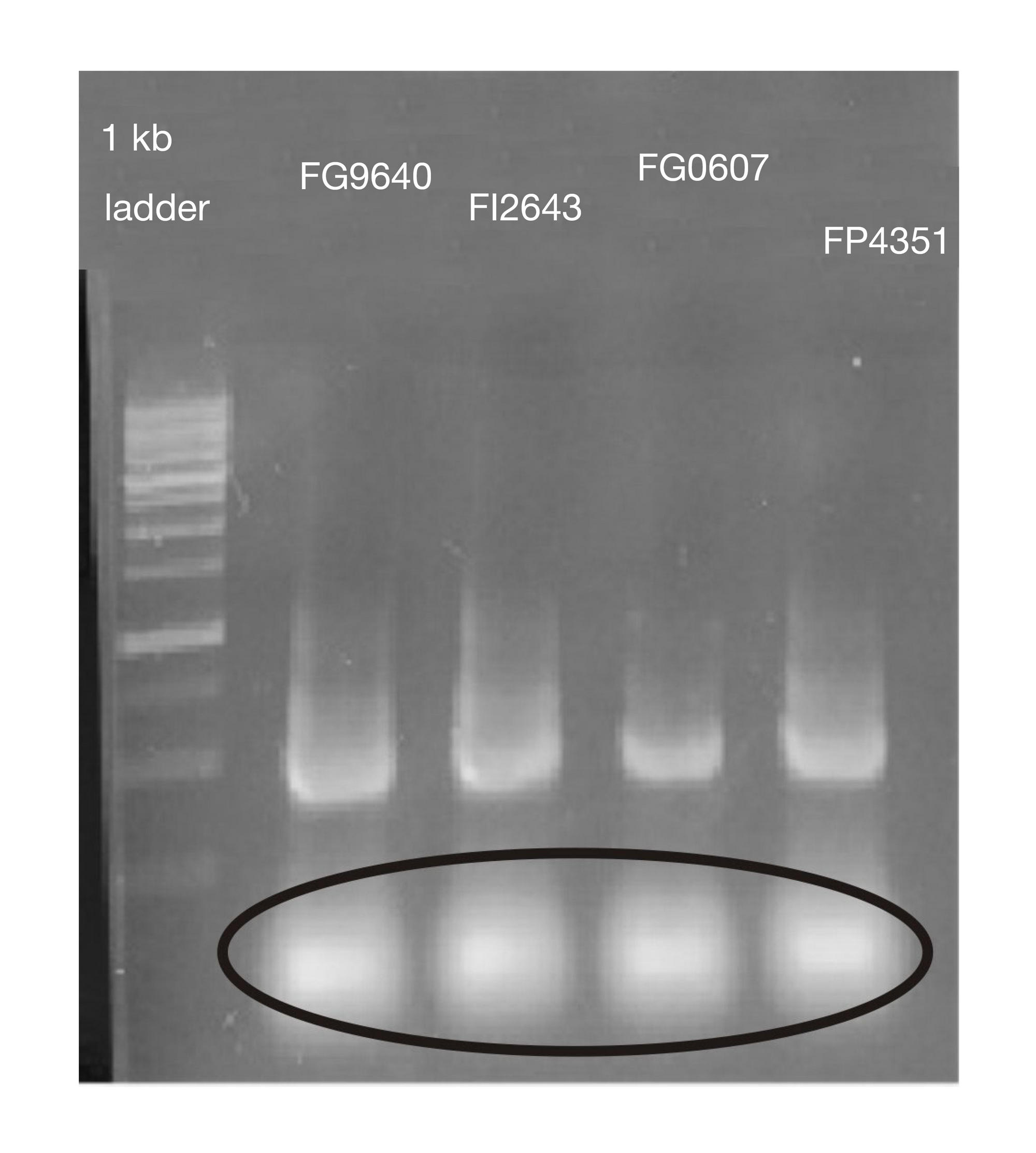
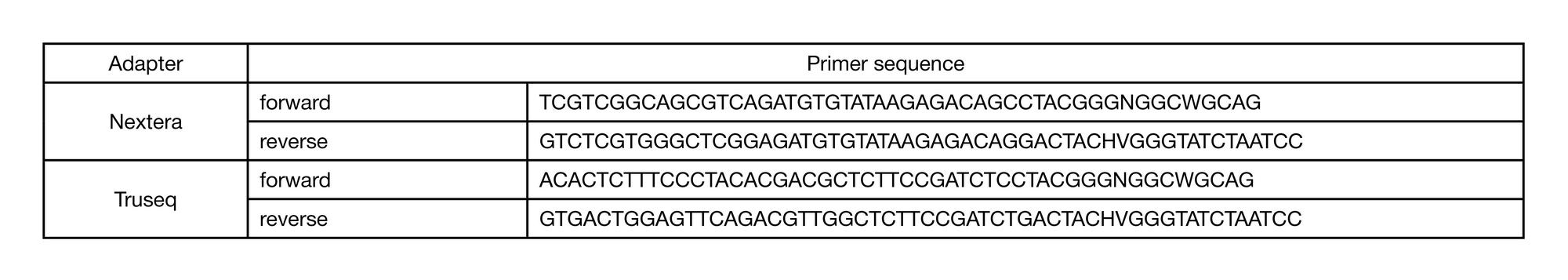
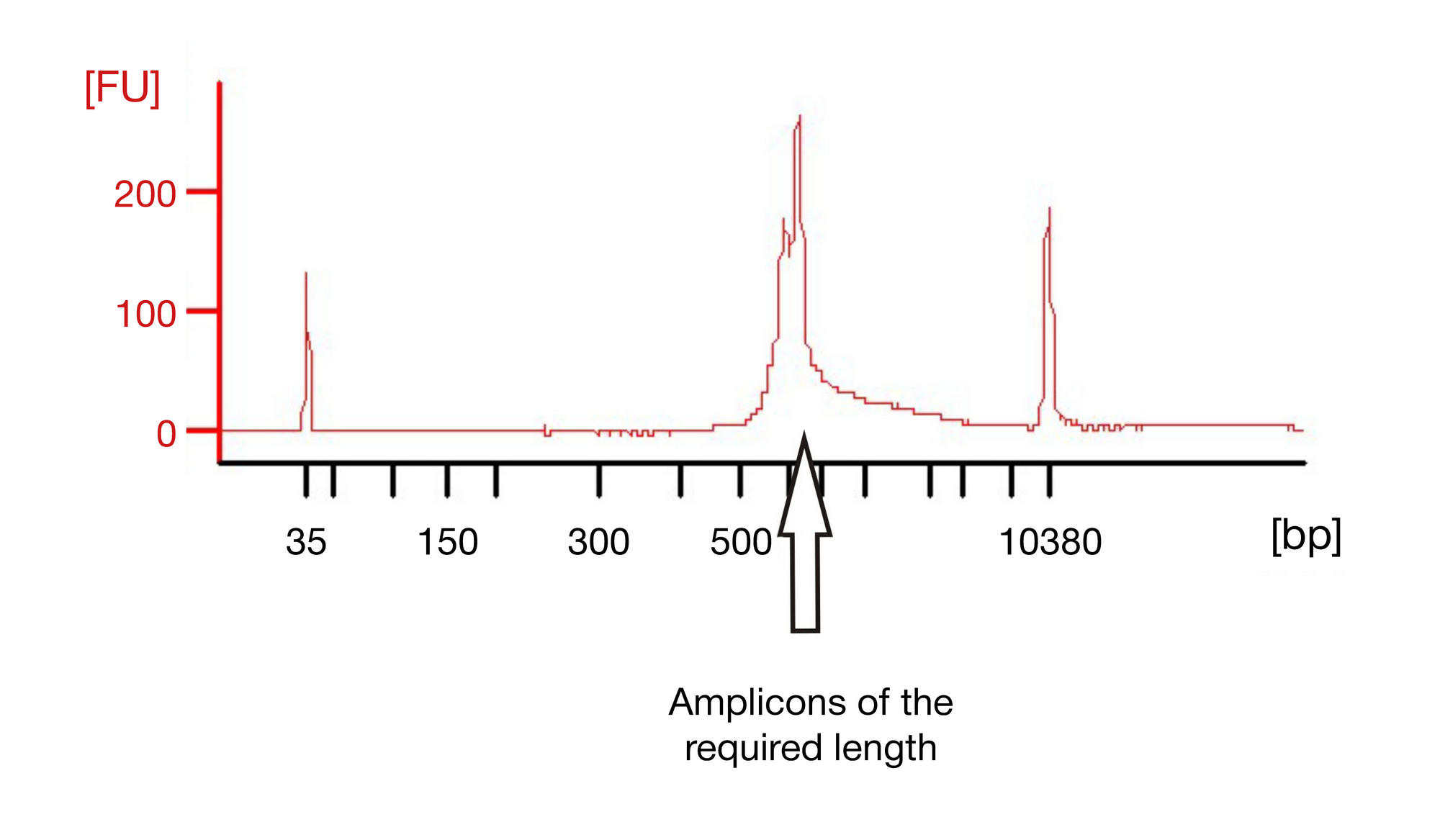
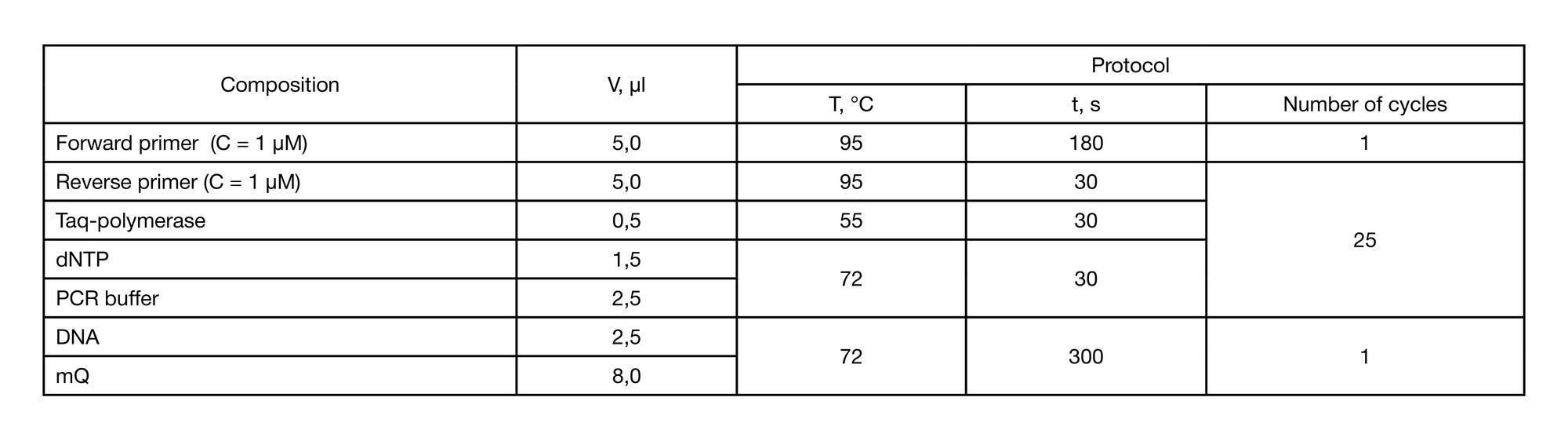
METHOD
Library preparation for metagenomic sequencing with Illumina
1 Genotek Inc., Moscow
2 Moscow South-West High School No. 1543, Moscow, Russia
Correspondence should be addressed: Anna Krasnenko
Nastavnicheskiy per. 17, str. 1, pod. 14, Moscow, 105120; moc.liamg@oknensarkanna
Acknowledgements: the authors thank Daria Plakhina and Ivan Stetsenok of Genotek for their help and Sergey Glagolev of Moscow South-West High School No. 1543 for his valuable advice and comments.
Contribution of the authors to this work: Krasnenko AYu, Eliseev AYu — analysis of literature, research planning and implementation, data analysis and interpretation; Borisevich DI, Tsukanov KYu — bioinformatic analysis; Davydova AI — drafting of a manuscript; Ilinsky VV — research planning, scientific advisor. All authors participated in editing of the manuscript.