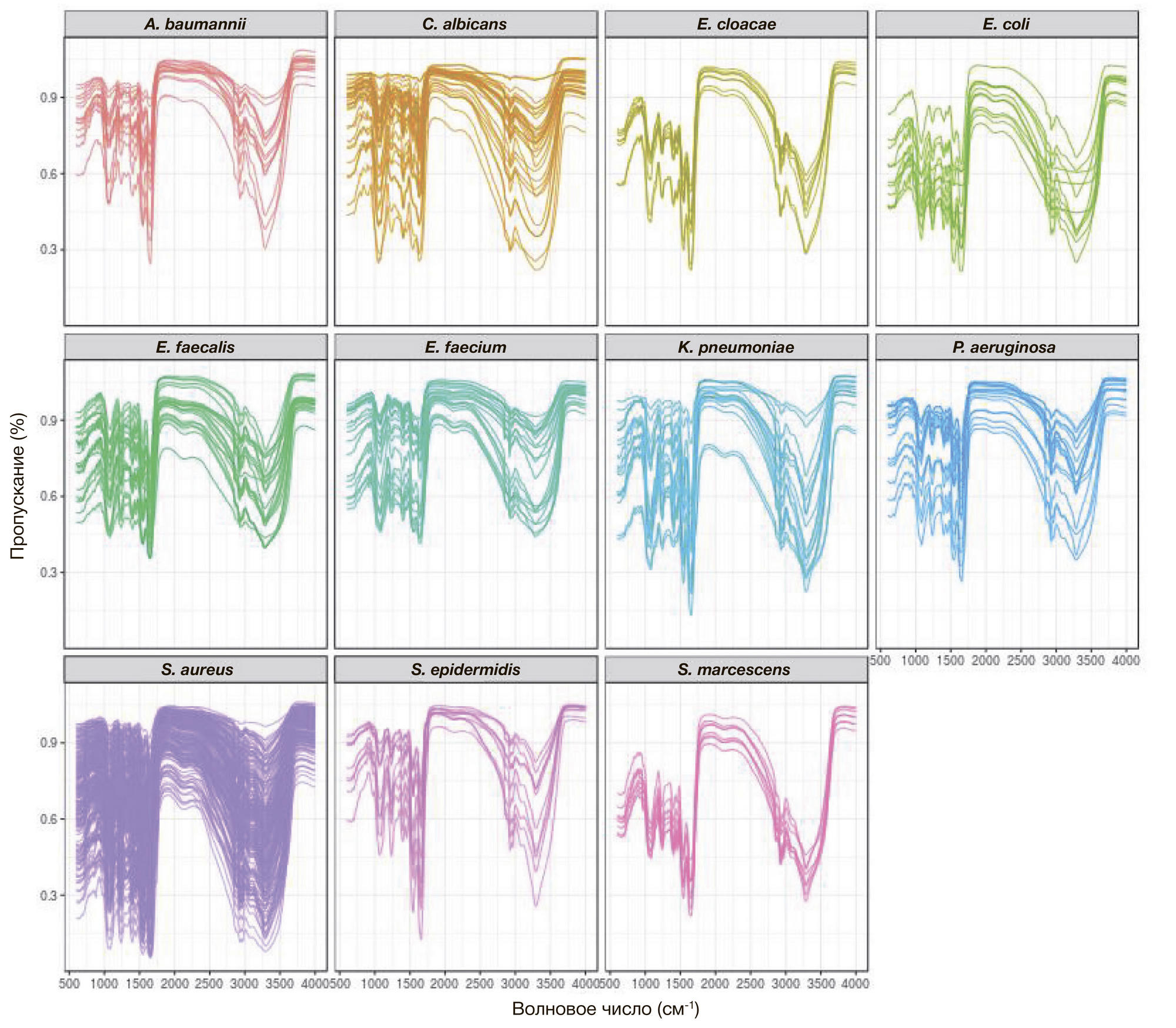
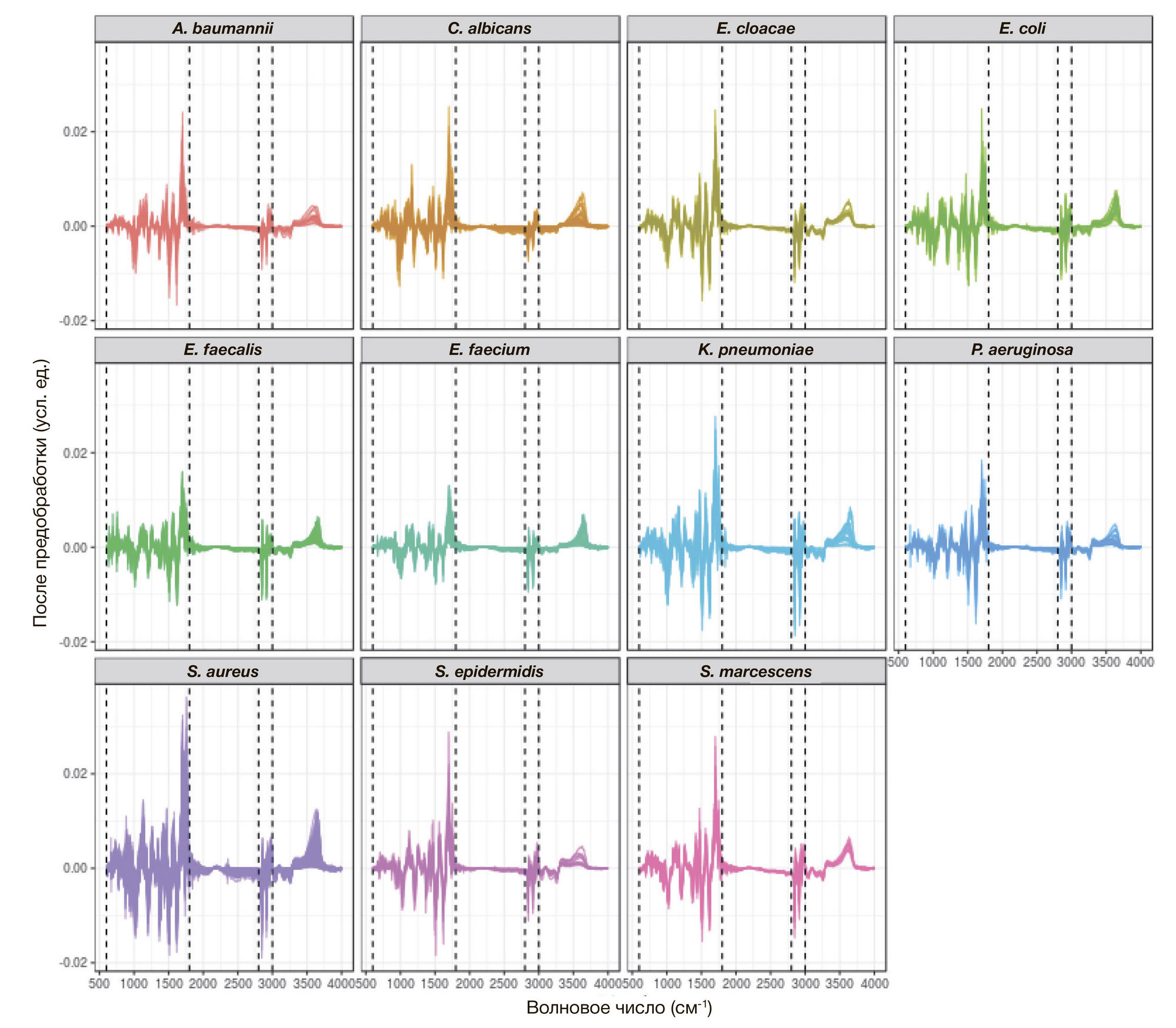
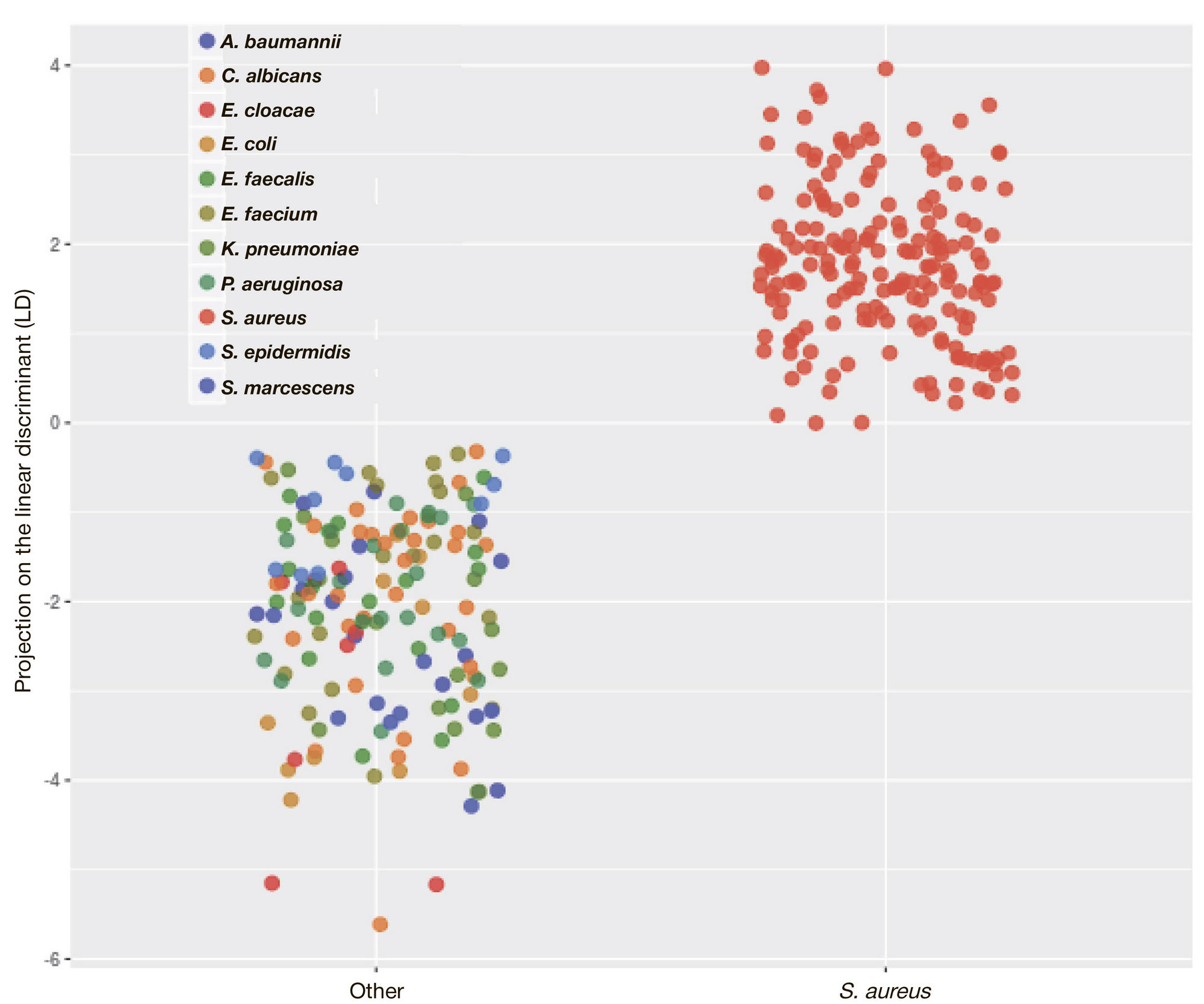
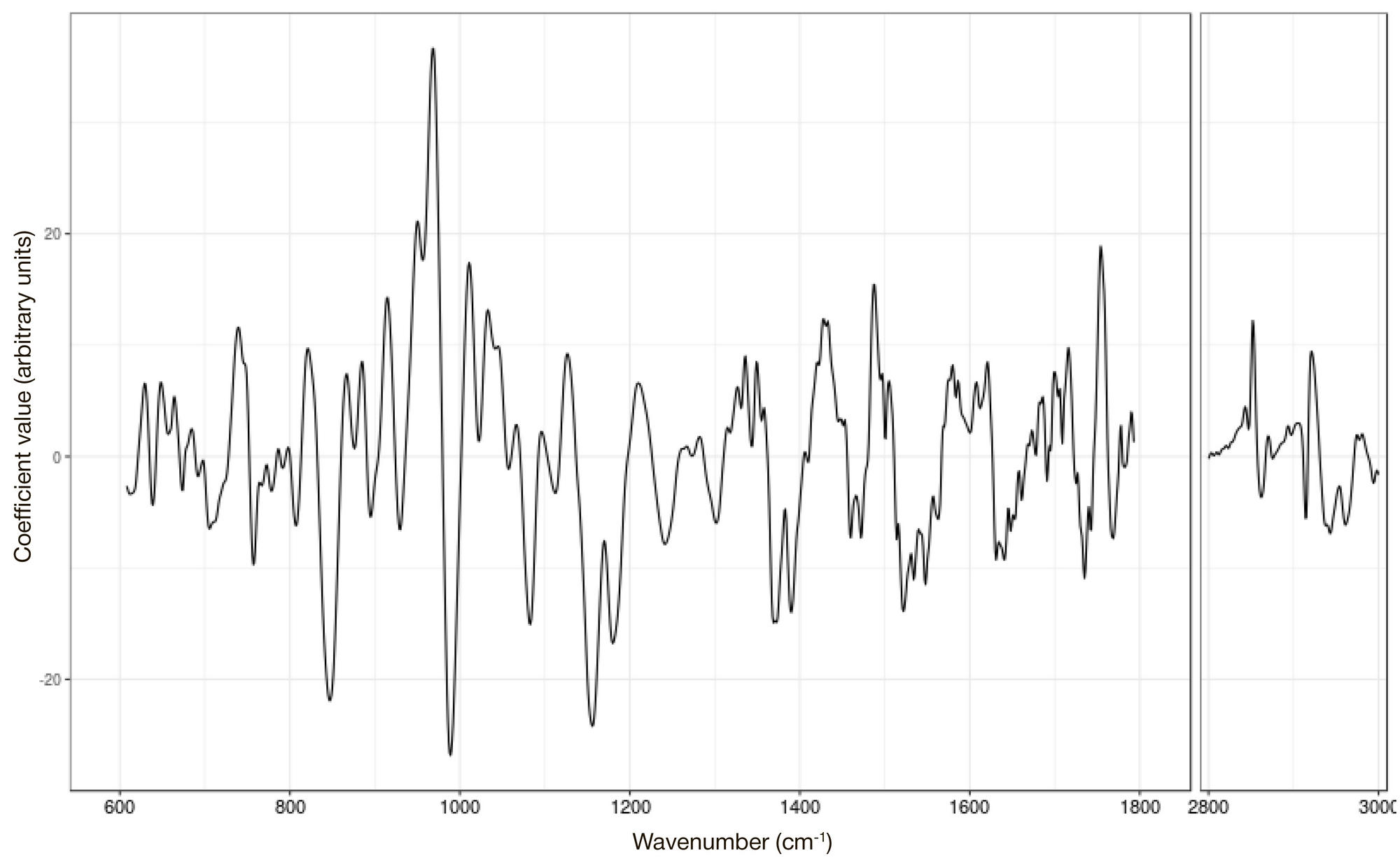
ORIGINAL RESEARCH
Identification of microorganisms by Fourier-transform infrared spectroscopy
1 Emanuel Institute of Biochemical Physics of the Russian Academy of Sciences, Moscow, Russia
2 Bakulev National Medical Research Center of Cardiovascular Surgery, Moscow
3 Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Moscow, Russia
4 Vavilov Institute of General Genetics, Russian Academy of Sciences, Moscow, Russia
5 Skryabin Moscow State Academy of Veterinary Medicine and Biotechnology, Moscow
Correspondence should be addressed: Alexei B. Shevelev
Kosygina 4, Moscow, 119334; moc.liamtoh@a_levehs