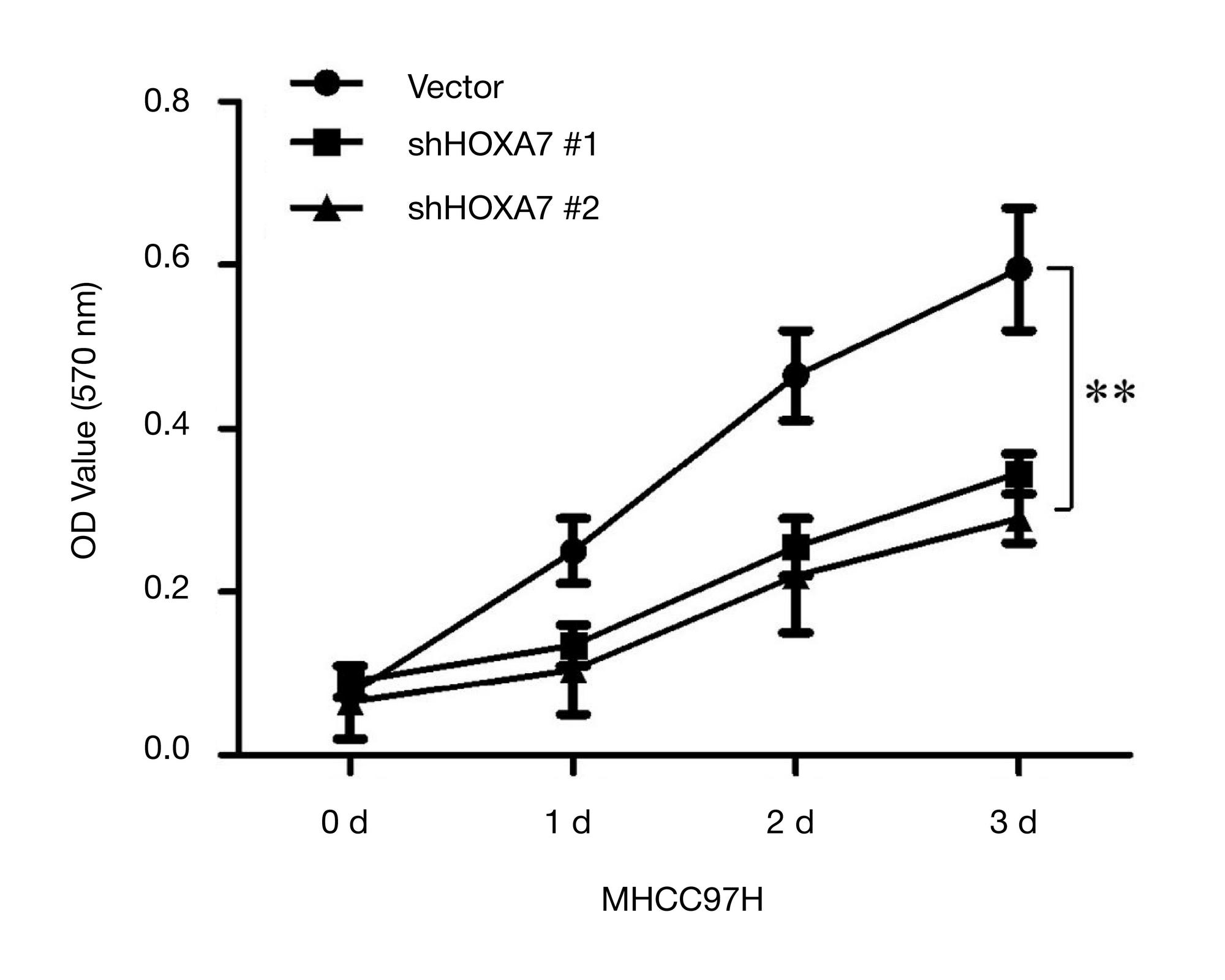
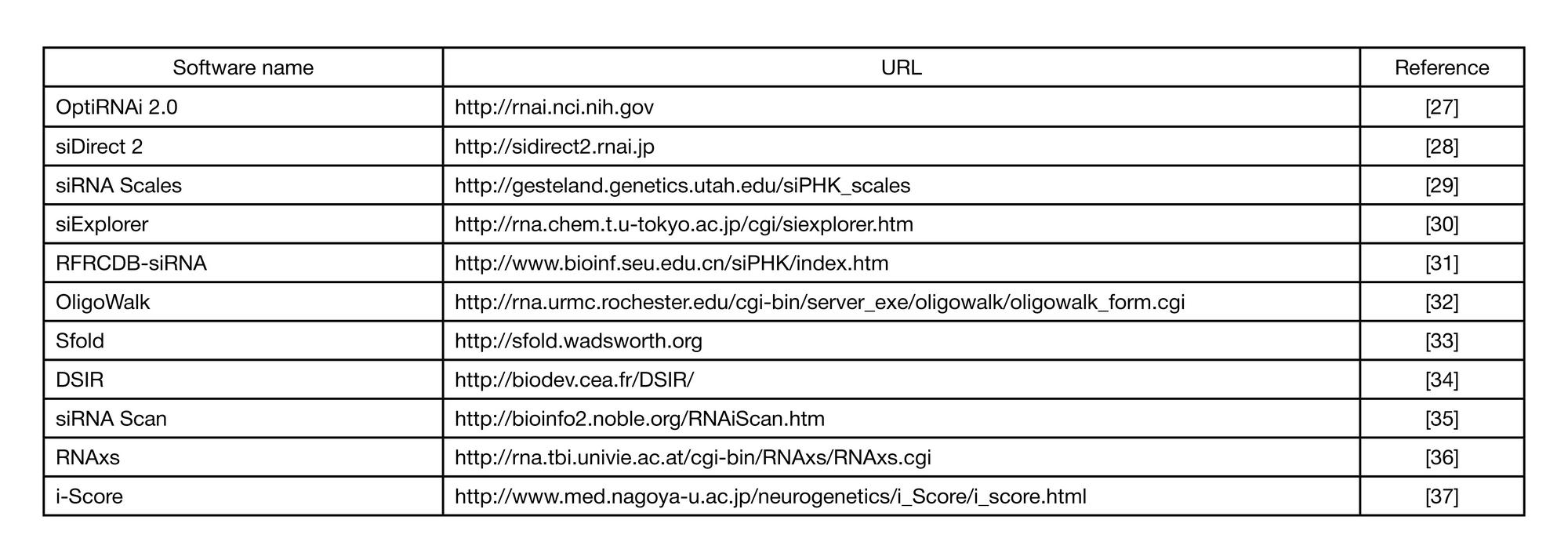
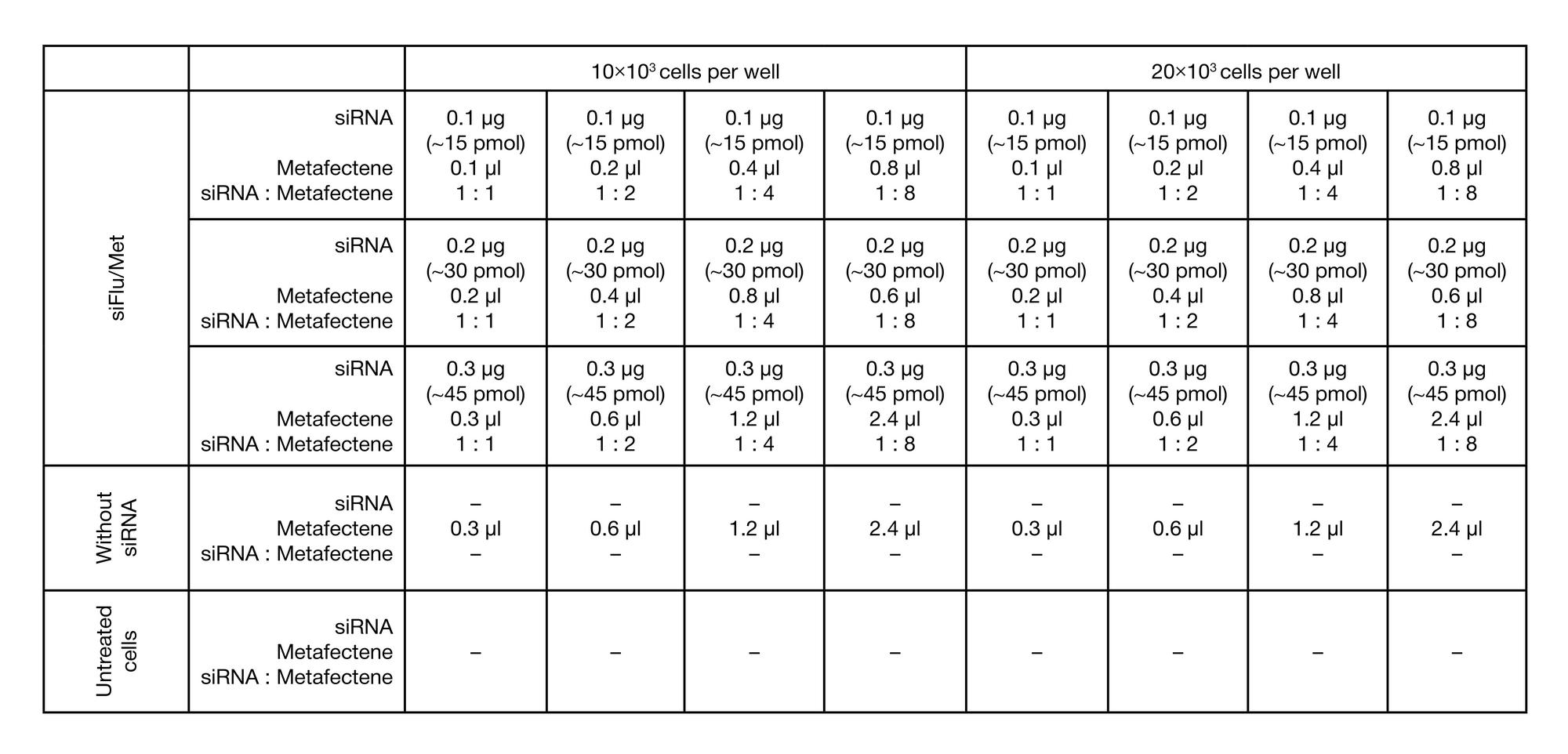
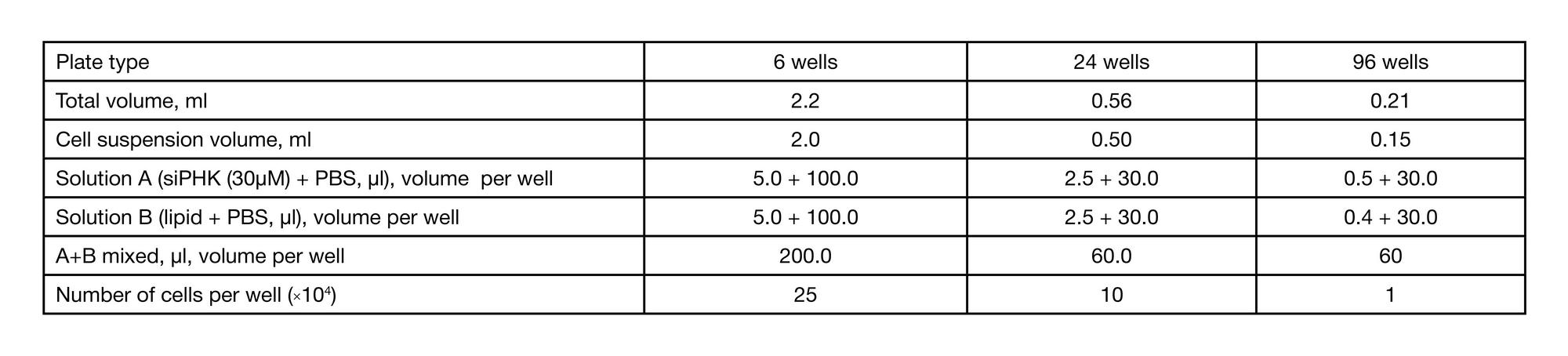
METHOD
siRNA-mediated gene silencing
1 Laboratory of Functional Genomics,Research Centre of Medical Genetics, Moscow, Russia
2 Laboratory of Medical and Genetic Technologies, Department of Basic Research of Research Institute for Medicine and Dentistry,Yevdokimov Moscow State University of Medicine and Dentistry, Moscow, Russia
3 Genomic Functional Analysis Laboratory,Moscow Institute of Physics and Technology (State University), Dolgoprudny, Russia
Correspondence should be addressed: Mikhail Skoblov
ul. Moskvorechie, d. 1, Moscow, Russia, 115478; moc.liamg@volboksm
Contribution of the authors to this work: Vyakhirava JV — analysis of literature, research planning, data collection, analysis and interpretation, drafting of a manuscript; Filatova AYu — analysis of literature, data collection, analysis and interpretation, drafting of a manuscript; Krivosheeva IA — analysis of literature, data collection, analysis and interpretation, drafting of a manuscript; Skoblov MYu — drafting of a manuscript. All authors participated in editing of the manuscript.