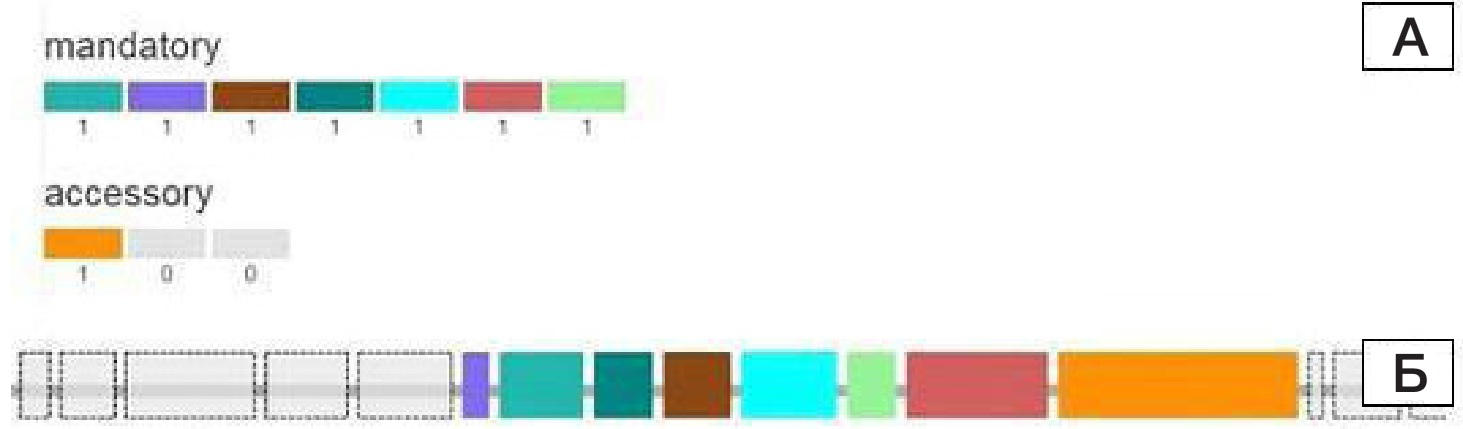
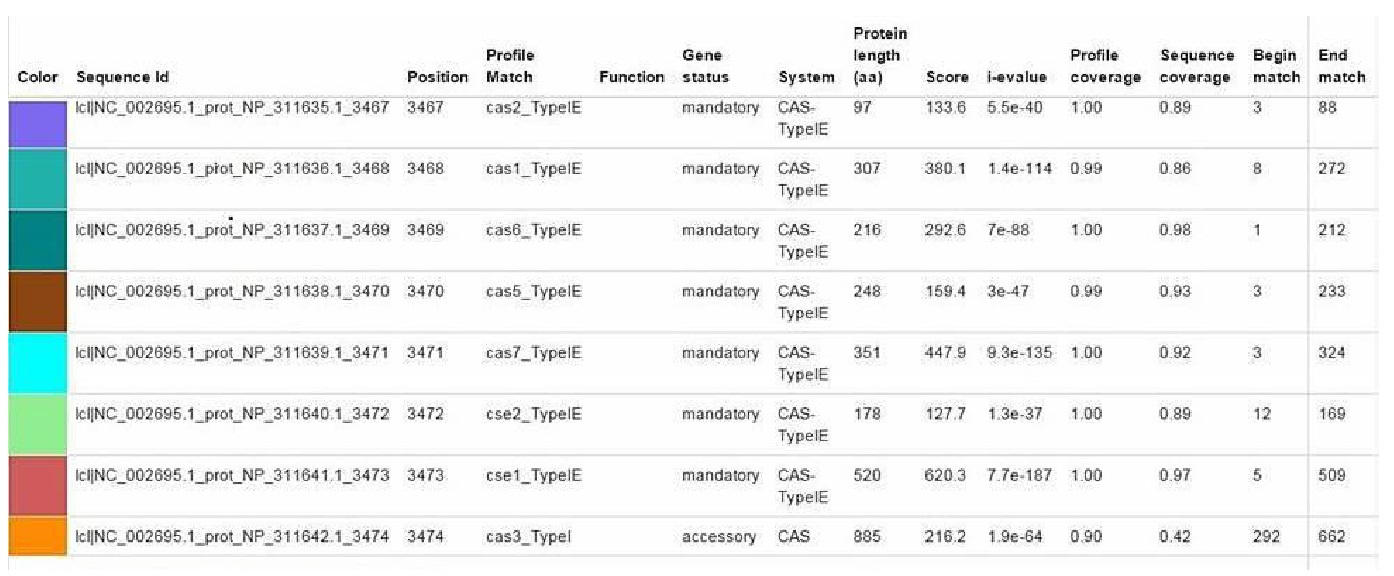
ISSN Print 2500–1094
ISSN Online 2542–1204
Bulletin of RSMU
BIOMEDICAL JOURNAL OF PIROGOV UNIVERSITY (MOSCOW, RUSSIA)

1 Scientific Center for Family Health and Human Reproduction Problems, Irkutsk
2 Research Institute for Biomedical Technologies of Irkutsk State Medical University, Irkutsk
Correspondence should be addressed: Elena Ivanova
Timiryazeva 16, Irkutsk, 66400; moc.liamg@mei.avonavi