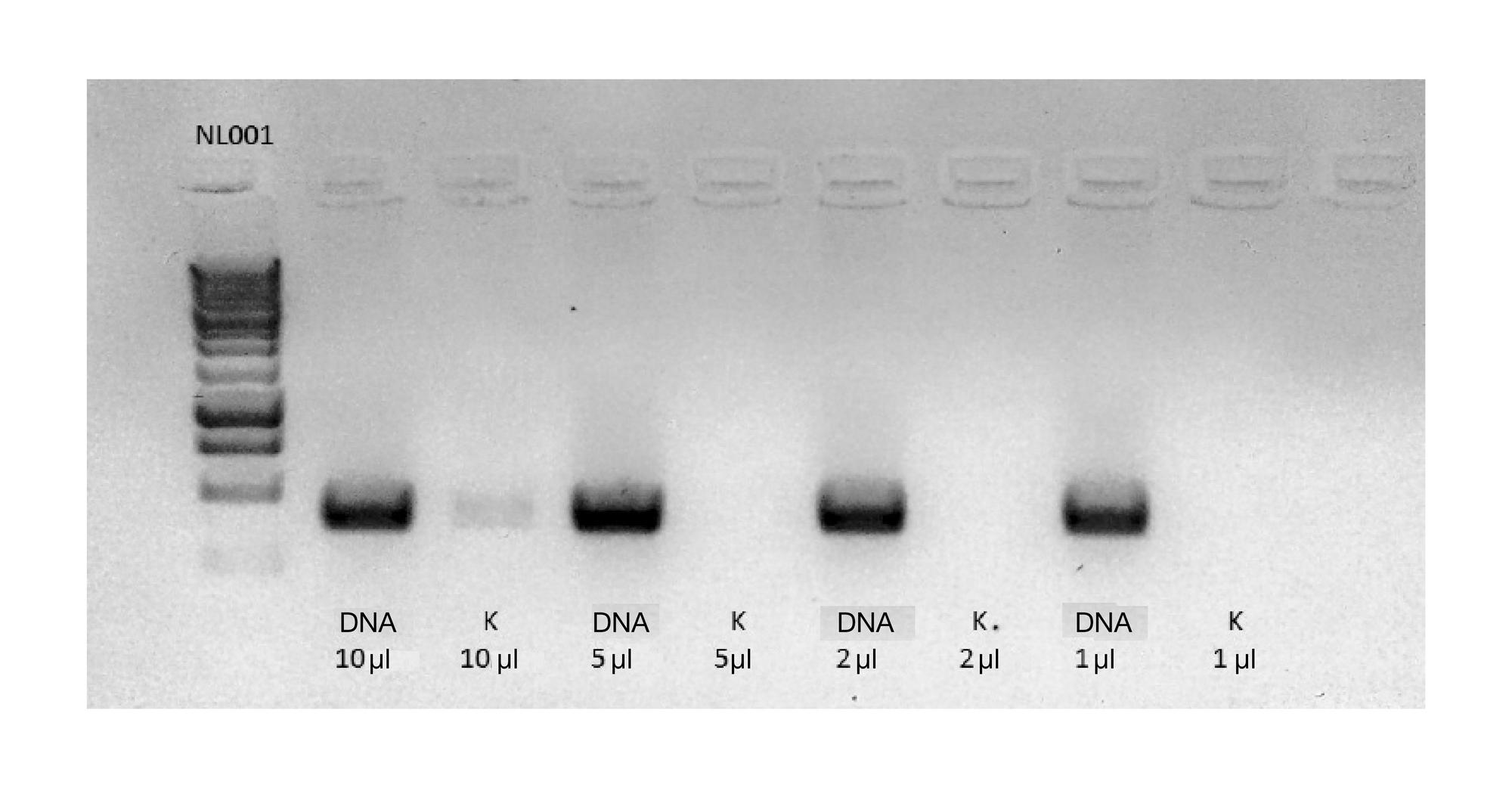
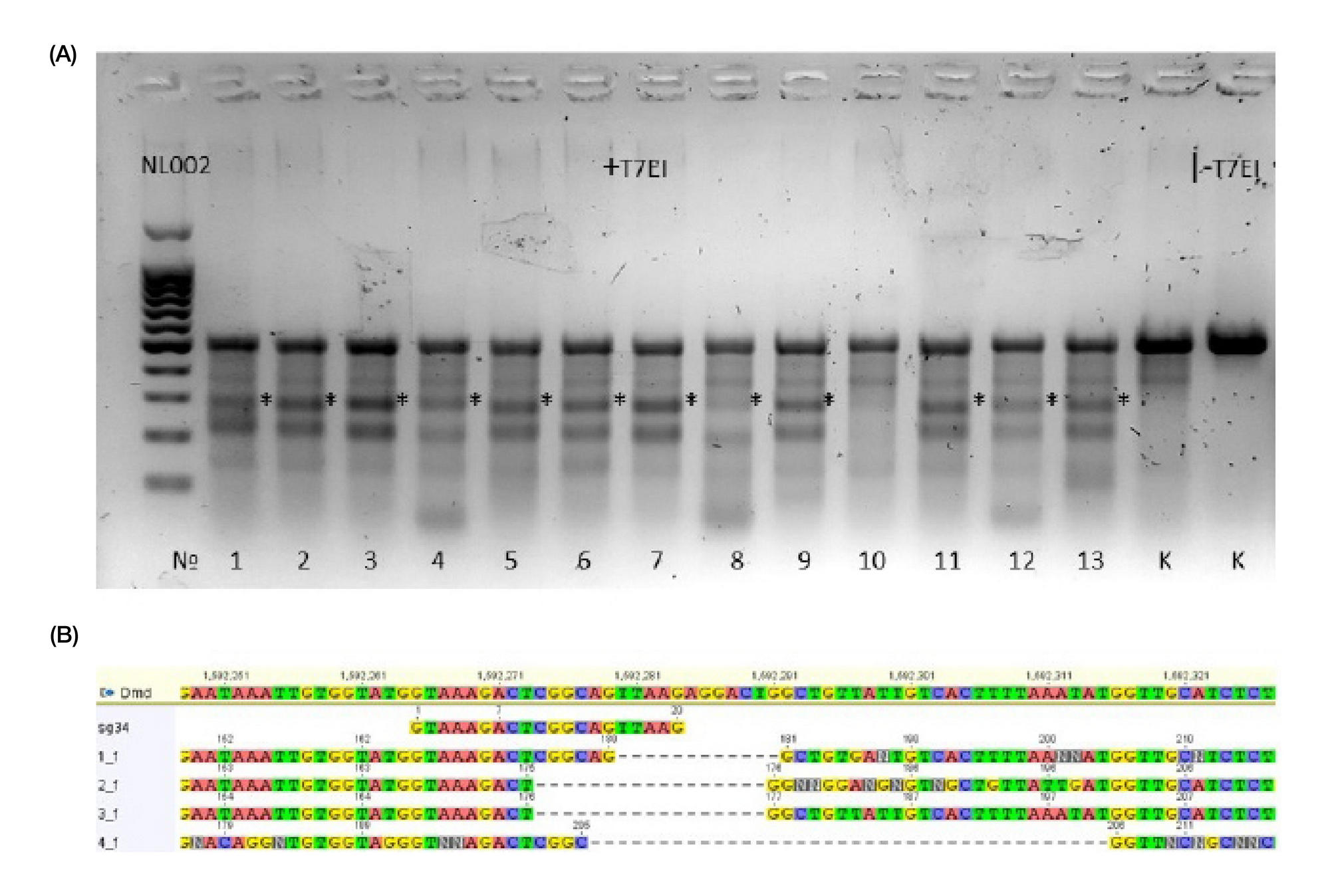
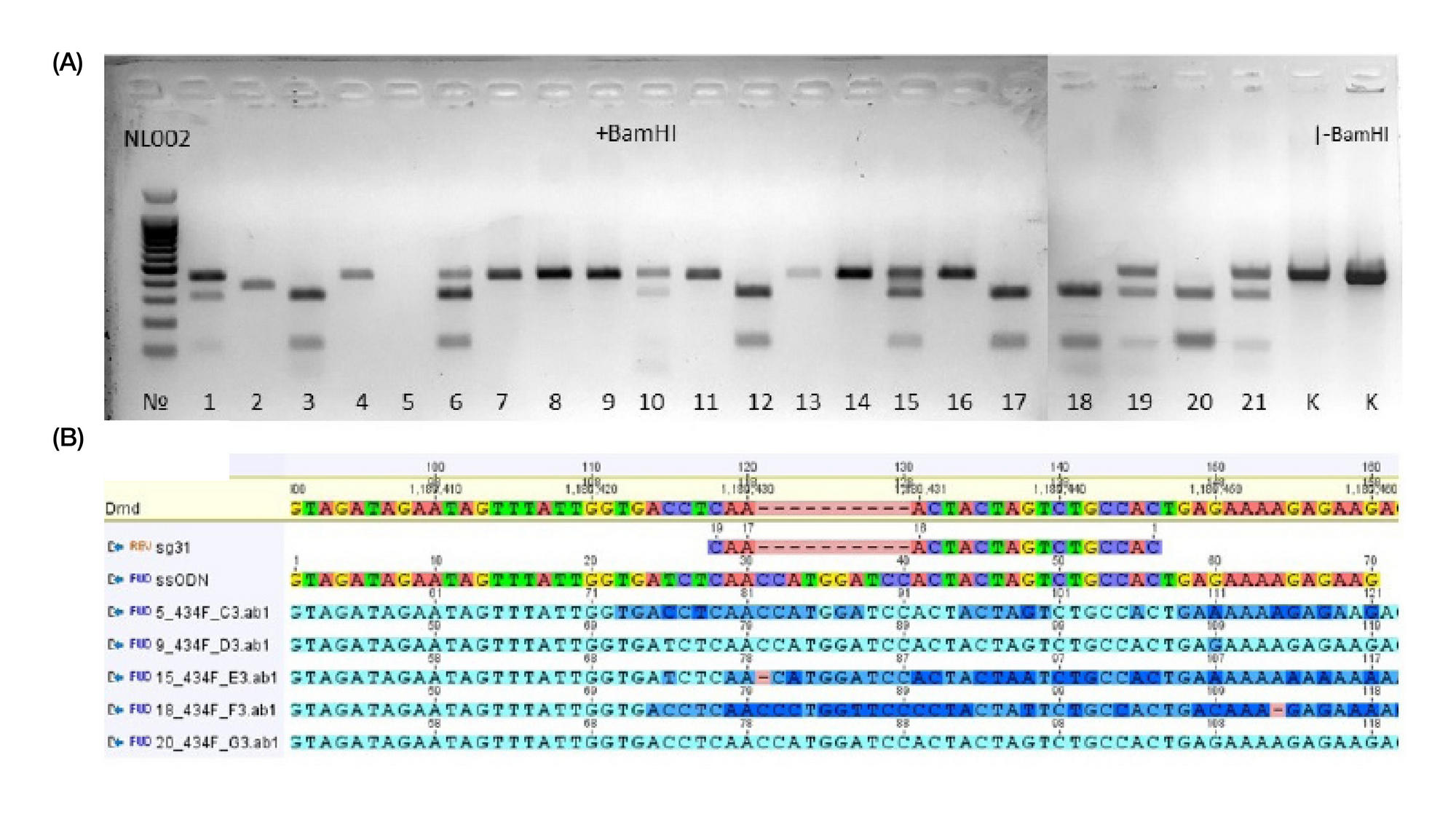
METHOD
Modification of the method for analysis of genome editing results using CRISPR/Cas9 system on preimplantation mouse embryos
1 Marlin Biotech, Moscow, Russia
2 Institute of Gene Biology, Russian Academy of Sciences, Moscow, Russia
3 Department of Neurology, Neurosurgery and Medical Genetics, Medical Faculty,
Pirogov Russian National Research Medical University, Moscow, Russia
Correspondence should be addressed: Tatiana Dimitrieva
ul. Vavilova, d. 34/5, Moscow, Russia, 119334; moc.liamg@at.aveirtimid
Acknowledgements: authors thank the Shared Resource Center of the Institute of Gene Biology of Russian Academy of Sciences for the equipment provided for this research.