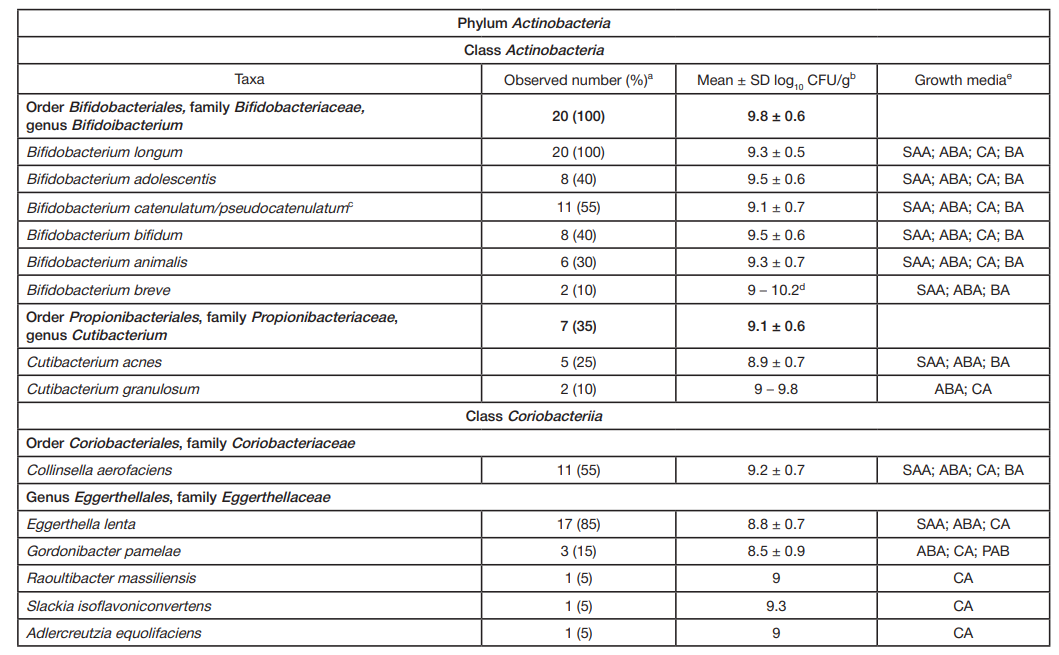
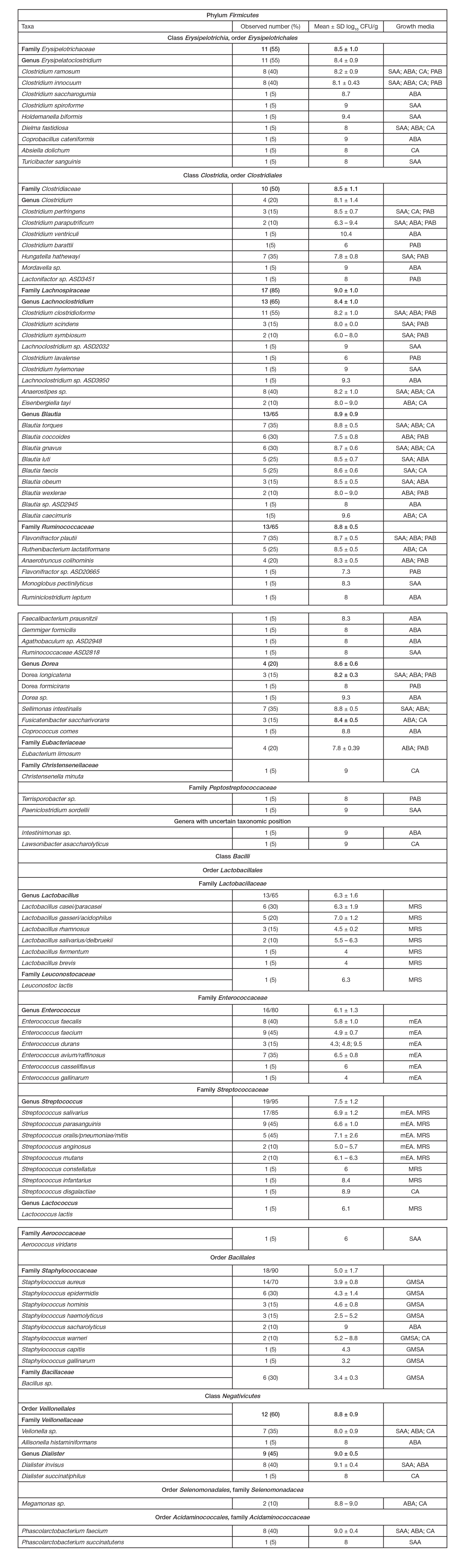
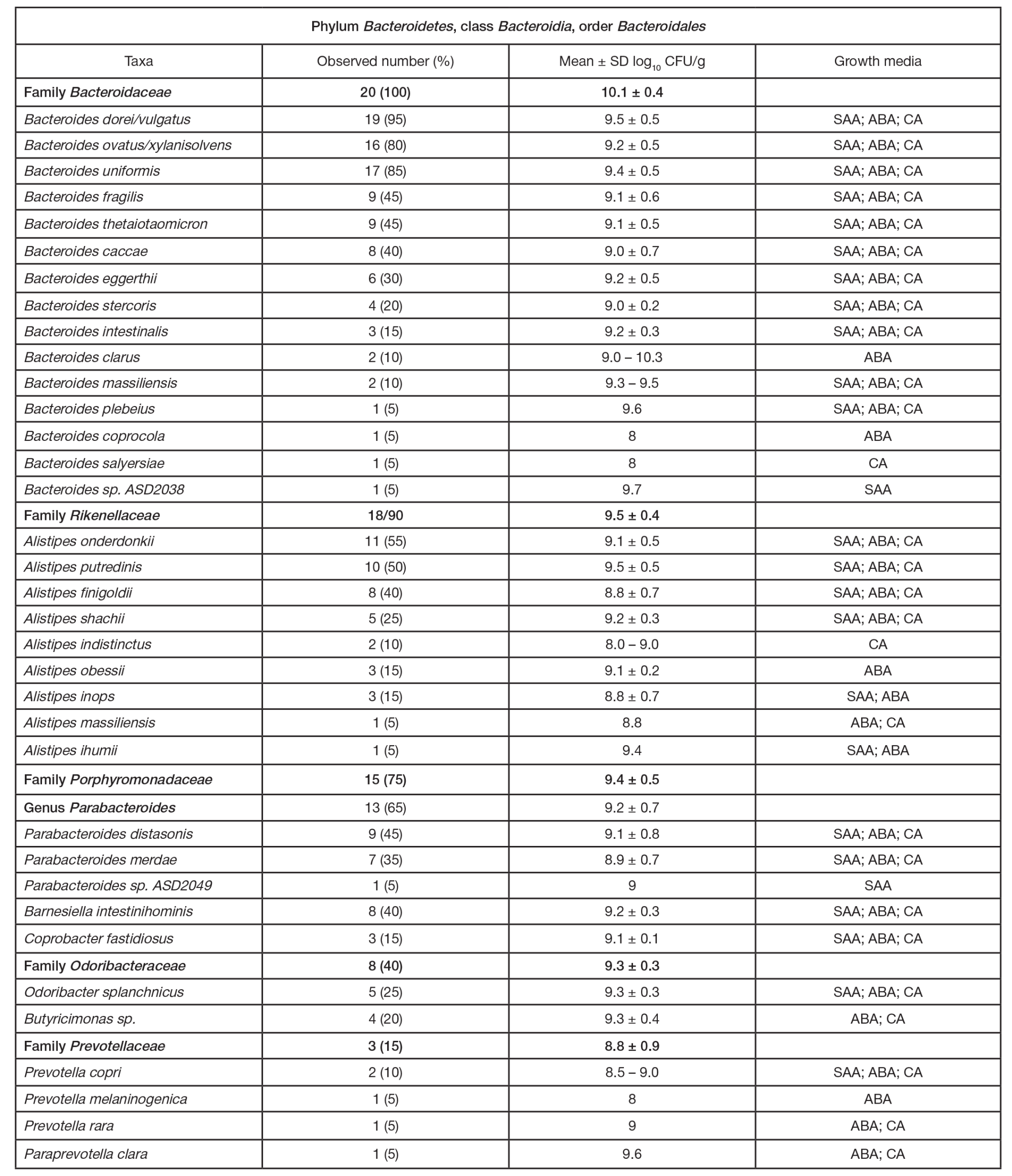
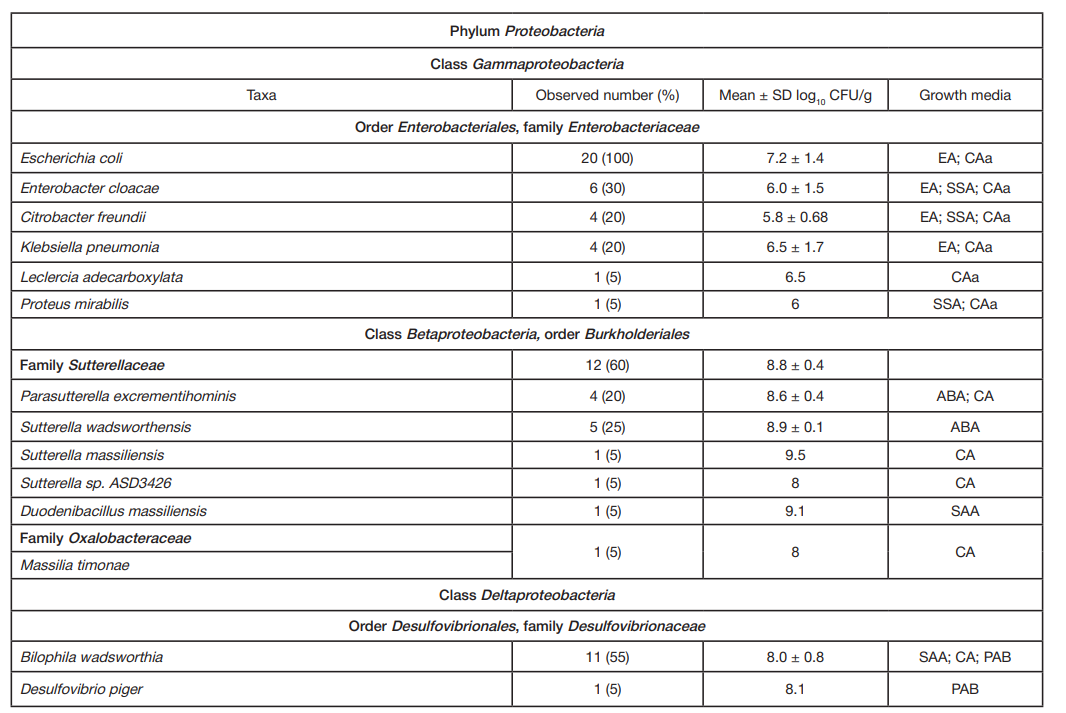
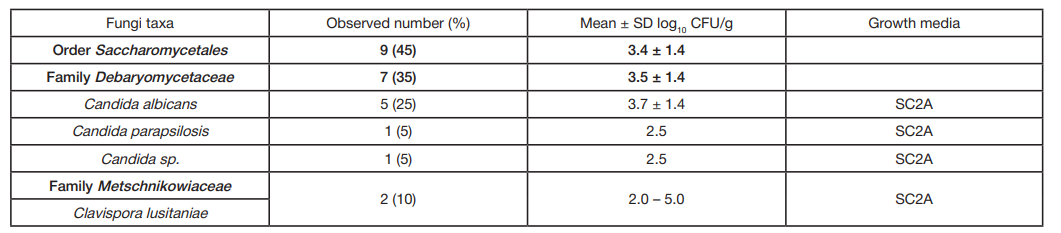
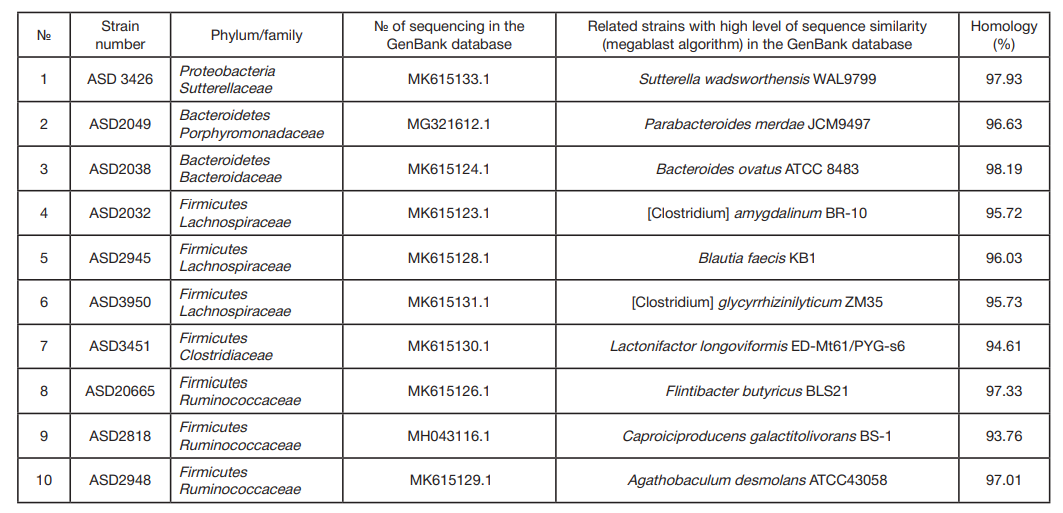
ORIGINAL RESEARCH
Application of culture-based, mass spectrometry and molecular methods to the study of gut microbiota in children
Pirogov Russian National Research Medical University, Moscow, Russia
Correspondence should be addressed: Boris A. Efimov
Ostrovityanova St. 1, Moscow, 117997; ur.liam@ab_vomife
Funding: the study was supported by Russian Science Foundation (research grant № 17-15-01488).
Author contribution: Efimov BA — research planning, literature analysis, screening children, specimen collection, microbiological research, mass spectrometry research, analysis and interpretation of data, preparing a draft manuscript; Chaplin AV — research planning, literature analysis, isolation of bacterial DNA, carrying out PCR, amplicons purification for sequencing, data analysis and interpretation, preparing a draft manuscript; Sokolova SR — research planning, literature analysis, specimen collection, microbiological research, isolation of bacterial DNA, carrying out PCR, amplicons purification for sequencing, data analysis and interpretation preparing a draft manuscript; Chernaia ZA — research planning, literature analysis, microbiological research, mass spectrometry research, data analysis and interpretation, preparing a draft manuscript; Pikina AP — research planning, literature analysis, microbiological research, mass spectrometry research, data analysis and interpretation, preparing a draft manuscript; Savilova AM — literature analysis, microbiological research, preparing a draft manuscript; Kafarskaya LI — research planning, literature analysis, screening children, microbiological research, analysis and interpretation of data, preparing a draft manuscript